Dicom Convert¶
Since in practice raw MRI data exported from an MR scanner consists of a large number of DICOM images storing slices and volumes of different sequences, by convention we convert the DICOM images to packed 3D or 4D NIFTI images before all processing steps. In BRANT, we use the dcm2nii from MRIcron/MRIcro to convert DICOM files into 4D NIfTI images by default and use wildcard characters to locate rs-fMRI image files. For the matched images, the First N timepoints removal is used to remove the first N frames that could be influenced by large motion or the instability of magnetic field.
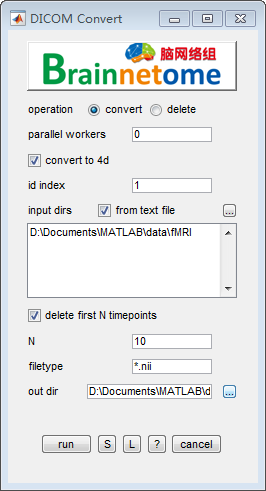
- operation: convert
- parallel workers: select how many workers to start a parallel work. The default is 0.
- convert to 4d: select to convert data to 4D format, otherwise to 3D.
- id index: identifier to find unique string for each subject.
- input dirs: directories can be input either using a .txt file or spm select window.
- delete first N timepoints: delete heading fMRI volumes.
- N: number of the heading timepoints to be deleted, the default is 10.
- filetype: wildcard to search wanted fMRI data.
- out dir: output directory for saving results.
- operation: delete
- id index: identifier to find unique string for each subject
- filetype: files in the filetype will be searched in input directories.
- 4D nifti files: if the input data is 4D, check this item. Otherwise uncheck.
- input dirs: directories can be input either using a
*.txtfile or spm select window. - delete first N timepoints: delete heading fMRI volumes.
- output fn: output filename.
- output to another directory: output data to another directory
- Buttons:
- S: Save parameters of the current panel to a
*.matfile. The*.matcan be further loaded for the panel or be used in a script processing. - L: Load parameters from
*.matfor the current panel. - ?: Help information.
- S: Save parameters of the current panel to a
- Reference:
- Rorden C, Brett M. Stereotaxic display of brain lesions. Behav Neurol 2000; 12(4): 191-200.