Extract Value¶
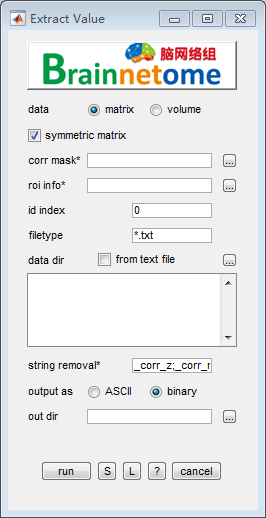
- data: matrix
- symmetric matrix: check to extract the upper right matrix’s elements, uncheck to extract all elements
- corr mask: e.g. a matrix mask used to find significant links instead of all links
- roi info*: a
.csvwith at least one column start with “label” - id index: identifier to find unique string for each subject
- filetype: filetype
- data dir: Input directories of matrices.
- data: volume
- roi file: ROI file used for extracting mean intensity in each roi tagged by number.
- roi info: labels of tagged ROIs. (optional)
- mask: could be whole brain mask or gray matter mask.
- id index: identifier to find unique string for each subject
- filetype: files in the filetype will be searched in input directories.
- 4D nifti files: if the input data is 4D, check this item. Otherwise uncheck.
- input dirs: directories can be input either using a
.txtfile or spm select window.
string removal*: optional. remove partial string from string parsed by id index.
output as: (only works for data type ‘matrix’) choose to output data matrix as ASCII(can be edited with text reader) or binary(more hard drive friendly with large matrix). if binary is chosen, you need to handle the matrix by:
fid = fopen('brant_extract_links.txt', 'rt'); outmat = fread(fid, sizeofmat, 'single'); fclose(fid);
out dir: output directory for saving results.
- Buttons:
- S: Save parameters of the current panel to a
*.matfile. The*.matcan be further loaded for the panel or be used in a script processing. - L: Load parameters from
*.matfor the current panel. - ?: Help information.
- S: Save parameters of the current panel to a